0. deepmd
使用案例路径:海康>A>A-6工作>deepkit使用案例
1.机器学习
1.安装:V3 版本
1.git clone -b v3 https://gitlab.com/Herbbb/torchdemo.git ****
2. source env_torchdemo (optional)
3.安装各种库,修改cmakelist/prifix.~前几行
**completer:**编译器
4.makedir build
cd build
cmake **../**cmakelist
make
5.编译和lammps的接口
把lammps的文件包复制过来,然后sh ~.sh build 18 (18是主节点核数,可以通过lscps 查看cpus的值获得)
2.训练势基本流程
1.从OUTCAR构建训练集,xsf格式,可直接在xcrystal 打开
sampleing.py
修改inv2
traindatapath = ./train-set*
batchsize = 1 和原子数相乘超过200
device = cpu
printstep = 100
hasattention = false 加不加注意力
训练
export OMP_NUM_THREADS=12 多线程
./
main.mp–train inv2 (是main.mp而不是lam.mpi)测试(修改inv2的测试部分)
./main.mp –eval inv2 保证能量相差几个meV,力相差几十meV,输出的单位是eV
注意:线程和核,48核是虚拟核,实际只有24物理核,提交一个6线程任务就会占据6个核训练机器学习势需要测试进程数,并不是越多越好,比如4远远好于48注意二:楼下集群做训练的速度太慢,最好不要用楼下集群训练
计算 (记得训练用的什么样的inv2计算就要用什么样子的inv2)。否则会报错,无法运行可以在训练过程中增加训练集,只要注意名字别和最开始的训练集重复
3.机器学习取样脚本
1. 按照目录来提取
import os
import sys
import time
import scipy.stats as st
import numpy as np
from sklearn.neighbors import KernelDensity
from matplotlib import pyplot
def timer(func):
def wrapper(*args, **kwargs):
start_time = time.time()
result = func(*args, **kwargs)
end_time = time.time()
print(f"Time elapsed: {end_time - start_time:.4f} seconds")
return result
return wrapper
def find_files(directory):
"""
递归查找一个目录下所有叫OUTCAR的文件,并返回这些文件的完整路径
"""
outcar_files = [] # 保存所有叫OUTCAR的文件的路径
for root, dirs, files in os.walk(directory):
for file in files:
if file == "OUTCAR":
outcar_files.append(os.path.join(root, file))
for dir in dirs:
dir_outcar_files = find_files(os.path.join(root, dir))
outcar_files.extend(dir_outcar_files)
# 将子目录添加到 dirs 中,以便递归处理
dirs[:] = [d for d in dirs if os.path.isdir(os.path.join(root, d))]
outcar_files = list(set(outcar_files))
return outcar_files
@timer
def read_vasp_xdatcar(file_path):
f = open(file=file_path)
f.readline()
f.readline()
lat = []
pos = []
# lattice
for _ in range(3):
lat.append(f.readline())
ele = f.readline()
npele = f.readline()
cnt = 0
tmp = None
while True:
line = f.readline()
if "configuration" in line:
cnt += 1
if tmp is not None:
pos.append(tmp)
tmp = []
continue
if not line:
pos.append(tmp)
break
tmp.append(line)
for conf_i in range(len(pos)):
for atom_i in range(len(pos[conf_i])):
pos[conf_i][atom_i] = pos[conf_i][atom_i].split()
pos[conf_i] = np.array(pos[conf_i]).astype(float)
ele = ele.split()
npele = npele.split()
npele = [int(i) for i in npele]
for i in range(len(lat)):
lat[i] = lat[i].split()
lat = np.array(lat).astype(float)
return cnt, ele, npele, lat, pos
def topn(arr, n):
indices = np.argpartition(arr, -n)[-n:]
values = arr[indices]
sort_indices = np.argsort(values)[::-1]
return values[sort_indices], indices[sort_indices]
def split_alp_num(string):
for i in range(len(string)):
if string[i].isdigit():
return [string[:i]], int(string[i:])
def write2my(file_path, ene_i, lat_i, ele_i, coo_i, foc_i):
lat_i = lat_i.reshape(3, 3)
coo_i = coo_i.reshape(-1, 3)
foc_i = foc_i.reshape(-1, 3)
with open(file_path, 'w') as file:
file.write(f"# total energy = {ene_i} eV\n\n")
file.write("CRYSTAL\n")
file.write("PRIMVEC\n")
for j in lat_i:
for k in j:
file.write(f'{k:20.8f}')
file.write('\n')
file.write("PRIMCOORD\n")
file.write(f"{len(coo_i)} 1\n")
for j in range(len(coo_i)):
file.write(f'{ele_i[j]:2}')
# coo
for k in coo_i[j]:
file.write(f"{k:20.8f}")
# force
for k in foc_i[j]:
file.write(f"{k:20.8f}")
file.write("\n")
pass
def direct2pos(lat, pos, direction=True):
"""
lat = [[Ax, Ay, Az],
[Bx, By, Bz],
[Cx, Cy, Cz]]
Pos = [n, 3]
:return: direct = [n, 3]
if direction=True
"""
if direction:
return pos @ lat
else:
return pos @ np.linalg.inv(lat)
class MyDistrib(st.rv_continuous):
def __init__(self, prdf):
super().__init__()
self.prdf = prdf
def _pdf(self, x, *args):
return self.prdf.evaluate(x)
class RejectSamp:
def __init__(self, ocr_name: str):
self.ocr_name = ocr_name # {XDATCAR path: OUTCAR path}
self.rng_min = 0
self.rng_max = None
self.rng_grid = 10000
self.rng = None
self.rng_intv = None
self.pdf = None
self.sum_intv = 10
self.envelope = None
self.samples = None
pass
def sampling(self, num):
nframe, natom, ele, ene, lat, pos, foc = self._anls_ocr()
print(nframe)
print(natom)
# print(ele)
print(ene.shape)
print(lat.shape)
print(pos.shape)
print(foc.shape)
self.rng_max = nframe
self.rng = np.linspace(self.rng_min, self.rng_max, self.rng_grid)
self.rng_intv = self.rng[1] - self.rng[0]
sd = np.sum((pos[1:] - pos[:-1]) ** 2, (1, 2))
nintv = sd.size // self.sum_intv
sum_sd = sd[:nintv * self.sum_intv].reshape(nintv, self.sum_intv).sum(1)
sum_sd_body = np.copy(sum_sd)
sum_sd_body[:sum_sd.size // 20] = sum_sd.min()
topn_val, topn_idx = topn(sum_sd_body, 100)
x_train = topn_idx * self.sum_intv
distrib = KernelDensity(kernel="gaussian", bandwidth=int(0.003 * nframe)).fit(x_train[..., None])
self.pdf = lambda x: np.exp(distrib.score_samples(np.reshape(x, [-1, 1])))
self.envelope = self.pdf(self.rng).max()
cnt = 0
iters = 0
res = np.array([])
while cnt < num:
u = np.random.uniform(self.rng_min, self.rng_max, num - cnt)
y = np.random.uniform(0, self.envelope, num - cnt)
pd = self.pdf(u)
res = np.append(res, u[pd > y])
res = res.astype(int)
res = np.unique(res)
cnt = len(res)
iters += 1
print(f"{iters:<3}{cnt:6}")
print("Total iters: ", iters)
self.samples = res
return self.samples.size, natom, ele, ene[self.samples], \
lat[self.samples], pos[self.samples], foc[self.samples]
def plot(self):
fig, ax = pyplot.subplots(figsize=(10, 6))
ax.hist(self.samples, bins=int(self.samples.size / 8), density=True, alpha=0.5)
ax.plot(self.rng, self.pdf(self.rng), 'r-', lw=1)
ax.plot(self.samples, -0.01 * self.envelope - 0.03 * self.envelope * np.random.random(self.samples.size), "+k")
ax.set_ylim(-self.envelope * 0.05)
ax.set_title(self.ocr_name + " PDF")
return fig
@timer
def _anls_ocr(self):
pos_foc = []
ene = []
lat = []
ions_per = []
types = []
print(self.ocr_name)
with open(self.ocr_name, 'r') as file:
while True:
line = file.readline()
# eof break
if not line:
break
# ions per type
#if "POSCAR" in line:
# ions_per.append(line)
# continue
if "ions per type" in line:
ions_per.append(line)
if "VRHFIN" in line:
types.append(line)
# lat
if "BASIS-vectors" in line:
file.readline()
file.readline()
file.readline()
file.readline()
for i in range(3):
line = file.readline()
lat.append(line)
continue
# pos & force
if "TOTAL-FORCE (eV/Angst)" in line:
line = file.readline()
while True:
line = file.readline()
if not line:
break
if "------" in line:
break
pos_foc.append(line)
continue
# ene
if "energy without entropy" in line:
ene.append(line)
ene = np.array([i.split('=')[1].split()[0] for i in ene]).astype(np.float64)
# lat = np.array([i.split() for i in lat]).astype(np.float64)[:, :3].reshape(-1, 3, 3)
lat = np.array([[float(i[4:][:12]), float(i[4:][12:25]), float(i[4:][25:38])] for i in lat]).astype(np.float64)[:, :3].reshape(-1, 3, 3)
# ele pretreatment
types = [i.split('=')[1].split(':')[0].rstrip() for i in types]
ions_per = ions_per[0].split()[-4:]
ele = sum([[i] * int(j) for i, j in zip(types, ions_per)], [])
pos_foc = np.array([i.split() for i in pos_foc]).astype(np.float64)
nframe = len(ene)
natom = len(ele)
pos = pos_foc[:, :3][:nframe * natom].reshape(nframe, natom, 3)[:nframe]
foc = pos_foc[:, 3:][:nframe * natom].reshape(nframe, natom, 3)[:nframe]
lat = lat[:nframe]
return nframe, natom, ele, ene, lat, pos, foc
if __name__ == '__main__':
# ocr_files = find_files("/data/ljx/third/lizhao/LYSO/LYSO-src/old")
#训练结构路径
ocr_files = ["/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/1fs/2",
"/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/1fs/4",
"/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/2fs/2",
"/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/2fs/4"]
#ocr_files = ["/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/triple/1",
# "/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/triple/2"]
#训练集输出路径
out_path = "/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/"
if not os.path.exists(out_path):
os.mkdir(out_path)
out_data_path = os.path.join(out_path, "eval-set")
if not os.path.exists(out_data_path):
os.mkdir(out_data_path)
#每个数据取多少点,大概相乘在10000附近
smp_per_ocr = 1500
# mode = 'rand'
mode = 'kde'
# # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # #
# Be careful when changing the code below
print(f"total trajectories: {len(ocr_files)}")
if not os.path.exists(out_data_path):
os.mkdir(out_data_path)
xsf_tot_cnt = 0
if mode == 'kde':
for ocr_i in range(len(ocr_files)):
r = RejectSamp(ocr_files[ocr_i])
n_frame, n_atom, ele, ene, lat, pos, foc = r.sampling(smp_per_ocr)
fig = r.plot()
fig_path = os.path.join(out_path, str(ocr_i))
fig.savefig(fig_path)
for j in range(n_frame):
path_j = os.path.join(out_data_path, f'structure{str(smp_per_ocr * ocr_i + j).zfill(5)}.xsf')
ene_j = ene[j]
lat_j = lat[j]
ele_j = ele
coo_j = pos[j]
foc_j = foc[j]
write2my(path_j, ene_j, lat_j, ele_j, coo_j, foc_j)
xsf_tot_cnt += 1
2. 从单个目录下递归直接提取全部
注意root_dir和write_dir的位置,其中root_dir放到要提取的当前目录最好,并且使用相对路径,这样提取的文件名称比较短
而write_dir则需要新建一个目录来存储提取的结构
import os
from ase.io import read
from ase import Atoms
def write2my(file_path, ene_i, lat_i, ele_i, coo_i, foc_i, vir_i=None):
lat_i = lat_i.reshape(3, 3)
coo_i = coo_i.reshape(-1, 3)
foc_i = foc_i.reshape(-1, 3)
with open(file_path, 'w') as file:
file.write(f"# total energy = {ene_i} eV\n\n")
if vir_i is not None:
file.write(f"VIRIAL\n")
for i in vir_i:
file.write(f'{i:20.8f}')
file.write("\n")
file.write("CRYSTAL\n")
file.write("PRIMVEC\n")
for j in lat_i:
for k in j:
file.write(f'{k:20.8f}')
file.write('\n')
file.write("PRIMCOORD\n")
file.write(f"{len(coo_i)} 1\n")
for j in range(len(coo_i)):
file.write(f'{ele_i[j]:2}')
# coo
for k in coo_i[j]:
file.write(f"{k:20.8f}")
# force
for k in foc_i[j]:
file.write(f"{k:20.8f}")
file.write("\n")
pass
def read_from_ase_atoms(atoms: Atoms):
ene = atoms.get_potential_energy()
lat = atoms.get_cell()
pos = atoms.get_positions()
foc = atoms.get_forces()
try:
sts = atoms.get_stress()
xx, yy, zz, yz, xz, xy = - sts * atoms.get_volume()
vir = np.array(
[[xx, xy, xz],
[xy, yy, yz],
[xz, yz, zz]]).reshape(-1)
except:
vir = None
ele = atoms.get_chemical_symbols()
return ene, lat, pos, foc, vir, ele
def find_files(directory, filename):
result = []
for root, dirs, files in os.walk(directory):
for file in files:
if file == filename:
result.append(os.path.join(root, file))
return result
if __name__ == '__main__':
root_dir = "./"
outcar_dir = find_files(root_dir, "OUTCAR")
write_dir = "/work/home/xieyu/workplace/liz/2-Li-Y-Si-O-600K/B-升温/data_xsf"
for i in outcar_dir:
xsf_head = '_'.join(i.split('.')[-1].split('/'))
print(xsf_head)
datas = read(i, index=":")
for n, i in enumerate(datas):
ene, lat, pos, foc, vir, ele = read_from_ase_atoms(i)
write2my(
os.path.join(write_dir, f"{xsf_head}_struct{str(n).zfill(6)}.xsf"),
ene_i=ene,
lat_i=lat,
ele_i=ele,
coo_i=pos,
foc_i=foc,
vir_i=vir)
4.遇到的报错、问题
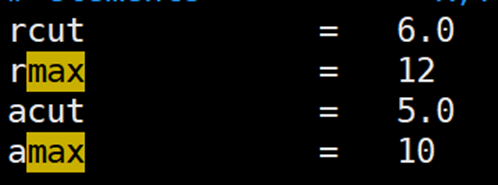
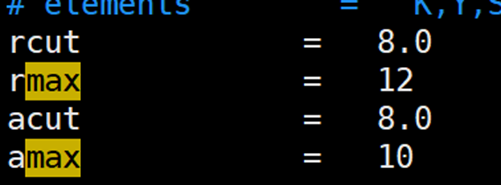
晶胞中某种原子的个数少,导致小的截断半径探索不到足够的数据,需要增加截断半径
解决方法:增加rcut和acut

找师弟
取样脚本报错,原因是中文名称路径
5.机器学习训练增加对压力的训练
- 在从OUTCAR中提取出的
- 在inv2中vpref的值不能为0,可以设置为与epref相等。
6. inv2中的元素类型的顺序
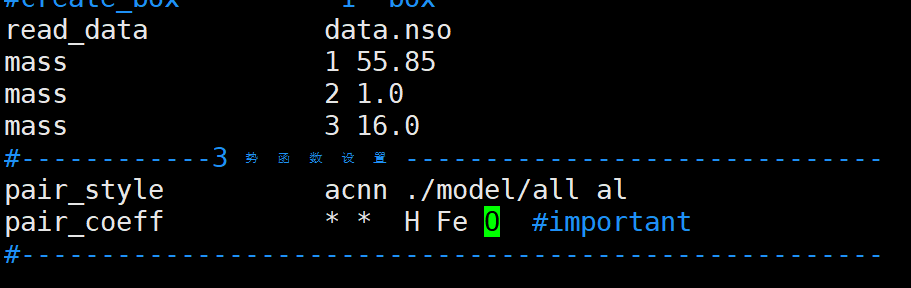
pair_coeff应该和初始的data.nso或者初始的POSCAR的原子顺序相同,而不是按照原子序数的大小变化
2. 主动学习机器学习
如果给主动学习一个很大的初始训练集,会降低主动学习的速度,原因是主动学习需要遍历很多的结构
初始训练集太少会导致一些较少的原子检测不到,会报错
1.注意事项
1. 老版本
1 关于显卡
gtop查看显卡信息,注意别把显存提爆 占用显存大小和胞的大小有关,和数据集数量无关
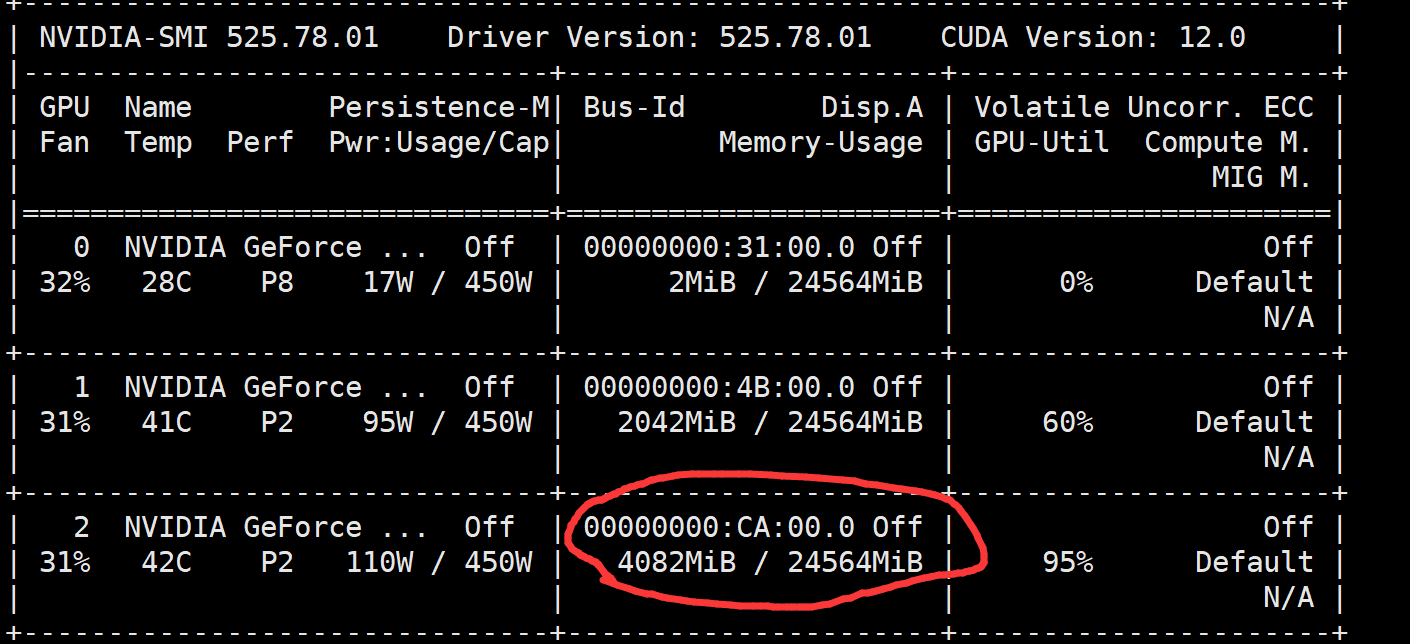
2 关于主动学习
对每个主动学习任务,需要在集群处建立新的文件夹,来进行自洽计算,避免多个任务进行时发生‘哈希碰撞’
修改下面文件中的路径


cal.slurm里提任务的名称要不同,因为是依据任务名称来回收任务的
记得激活python3环境(不然不写新找到的数据集,现在修复了)
修改使用哪个卡
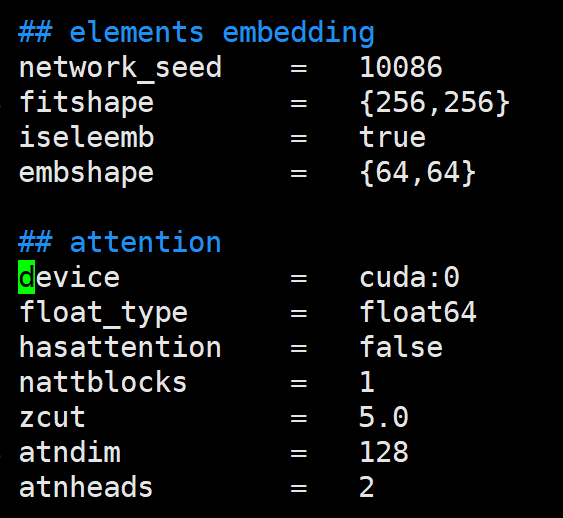
可以把多个任务提交到一块卡上面
如果是单独只训练需要把restart修改为false
不能把poscar放到fp里面,否则会反复lable 一个结构
任务输出的log文件要统一命名 3-md-log(lammps分子动力学输出)
2-train-log(训练输出)
4-select-log (选择输出)
vasp.log vasp输出
训练的圈数不需要太多 3000-5000
初始训练集的数目100左右
acnn的路径(sub-train.sh sub-select.sh)要正确
inv2 里rcut等
注意sub-md.sh和sub.slurm里面的任务名称不但不同任务要不同,而且要与原来集群上的任务名称不同
waiting remote jobs: 1 含义是有一个等待回收的任务
lr是学习率,学习率的值减小,说明模型的变化量减小
inv2中 select_thr 控制挑选结构的下限阈值。目的是挑选出结构差异性大的结构,增大sub-select.log中target的值(一个矩阵的行列式,不同行差异性越大行列式越大)。初期因为跑的少,很多相似结构,所以阈值要高点,不能什么都加进去,后期跑的多了,可以把阈值调低点,增加多样性,
现在推荐把select_thr设置为1.2
3.需要测试的事项
- 初始数据集的大小多少更合适
- 训练的圈数需要随着主动学习次数的增加而变化,多少圈比较合适
- 小包跑多少算合理了
- 主动学习过程中,哪些是占资源多的,哪些是花费时间长的
- nohup ./sub-train.sh > train.log 2>&1 &
- inv2 里卡的名称
- in.lmp 里质量、名称
- 给一个data.nlso文件
4.训练方式-增强采样
先用小胞,小圈数 训练到一定程度后(小胞可以跑足够长时间),用大胞、大圈数 目的是增加训练集
可以循环着提供大胞的扩胞方式
inv2 nbatch和savestep要联动修改
增加胞的大小应该可以减小代数
top -u lijx
跑升温过程训练,这样得到的势理论上可以跑不同的温度
增强采样:大晶胞、训练圈数增加、广的温度采样范围、长的模拟时间
5. 关于续算
1.没有跑到设置条件(没有跑足1ns…)
删除最后一代
在scheduling.sh里修改代数,不从0开始,而是输入刚刚删除的代的代号,这样,会自动从前一代读取最后的模型,进行训练。在提交任务后,新的一代内训练输出2-train.log中会出现提示(需要向下多翻)
例如,跑完是 28 29 30 ,那就删除30的文件夹,同时把scheduling.sh里改为30
提交任务应该注意 >> log 否则续算会把之前的log覆盖掉 nohup ./scheduling.sh >> log 2>&1 &
可以适当修改分子动力学里的晶胞尺寸、模拟时间、训练圈数和保存的步数、训练用到的卡
2.跑到预定条件,但是希望续算
正常训练结束后log会报没有输出oos的错误,可以通过看最后一代3md的输出log.lammps判断
不需要删除最后一代,只需要修改scheduling.sh为下一代开始,例如,出现了 28 29 30 那么scheduling.sh里修改为 31
需要把前一代的Ap_inv_*拷贝过来,因为没有select的步骤没有办法产生这些文件,但是分子动力学计算需要这些文件
Ap_inv_*用于主动学习中选择结构,如果用al的模式跑分子动力学,需要用到Ap_inv*来作为选择结构的标准,但是分子动力学过程并没有用到。用不用al跑出来的结果是一样的,差别是要不要获取结构可以不用al,也就不需要Ap_inv修改in.lmp里面的模拟晶胞尺寸、模拟时间、以及升温过程的温度(更广一些)、增加训练和保存的圈数、等等方式来增加采样
还有修改 restart_lr = 1e-4 学习率
学习率1e-7训练就差别不大了
inv2中 select_thr 调低 1.2
6 批量化杀掉当前文件夹内后台的任务(不想继续跑的时候)
# 获取用户 lijx的所有进程 ID
pgrep -u lijx > lijx_processes.txt
# 通过进程 ID 获取对应的进程所在目录
while read -r pid; do
ls -l /proc/"$pid"/cwd 2>/dev/null | grep -q "/public/lijx/work/Na-X-Si-O/2-onthefly/3-gpu" && echo "$ pid"
done < lijx_processes.txt > lijx_processes_in_test.txt
# 通过进程 ID 杀死位于 /home/lijx/test 及其子目录下的所有进程
while read -r pid; do
kill -9 "$pid"
done < lijx_processes_in_test.txt
for i in $(cat lijx_processes_in_test.txt);do llpid $i;done
2.集成化提任务方式
四个文件 1-convert_outcar_xsf.py 2-convert_POSCAR_data.py 3-makepot.py make_initial.sh
只需要配置make_initial.sh
#需要的文件:POSCAR OUTCAR sub.slurm(vasp) sub-md.sh(lammps)
#先切换到python3环境 conda activate
#set vasp_systerm
vasp_folder='"/work/home/liz/workspace/1-system-MD/5-Na-X-Si-O/md/1-Mo"'
md_duty_name='md-Mo'
vasp_label_duty_name='vasp-Mo'
#set GPU
GPU_number=0
#set md
cat > in.lmp << lzh
variable x index 1
variable y index 1
variable z index 1
variable ss equal 0.001
variable tdamp equal "v_ss*100"
variable dmsdx equal c_1[1]
variable dmsdy equal c_1[2]
variable dmsdz equal c_1[3]
variable dmsd equal c_1[4]
variable istep equal step
variable msdx format dmsdx %16.8e
variable msdy format dmsdy %16.8e
variable msdz format dmsdz %16.8e
variable msd format dmsd %16.8e
variable sstep format istep %-10g
units metal
atom_style atomic
boundary p p p
read_data ./data.nso
mass 1 22.990 #important
mass 2 95.94
mass 3 28.084
mass 4 15.999
replicate \$x \$y \$z
pair_style acnn ./model/all al
pair_coeff * * Na Mo Si O #important
thermo 1
thermo_style custom step time temp press pe lx ly lz ke
thermo_modify format 4 %20.15g
compute 1 all msd com yes
dump out all custom 100 A.lammpstrj id element xu yu zu fx fy fz
dump_modify out element 1 2 3 4
dump_modify out format 1 %4d
dump_modify out format 2 %4s
dump_modify out format float %16.8f
velocity all create 800.0 8728
timestep \${ss}
fix 1 all nvt temp 800 800 \${tdamp}
fix 2 all print 1 "\${sstep} \${msdx} \${msdy} \${msdz} \${msd}" screen no file msd.dat
run 1000000
lzh
#not always need changes
vasp_ip='"liz@59.72.114.246"'
vasp_port='"2222"'
command_line='"slurm"'
# set vasp_input
vasp_pot_PBE_Path='"/public/lijx/work/Na-X-Si-O/pot-makepot/potpaw_PBE/"'
cat > INCAR << liz
SYSTEM = Na-X-Si-O
ALGO = Normal
ISYM = 0
LREAL =Auto
PREC = Normal
EDIFF = 1e-5
ENCUT = 400
NELMIN = 4
NELM = 500
IBRION = -1
ISIF = 2
ISMEAR = 0
SIGMA = 0.05
NPAR = 6
LH5 = .TRUE.
liz
cat > KPOINTS << lizh
K-Spacing Value to Generate K-Mesh: 0.06
0
Gamma
1 1 1
0.0 0.0 0.0
lizh
sed -i "s|^base_pseudo_path.*|base_pseudo_path = $vasp_pot_PBE_Path|" 3-makepot.py
mv ../POSCAR ../resource/fp
mv ../sub.slurm ../resource/fp
mv ../sub-md.sh ../resource/md
mv ./INCAR ../resource/fp
mv ./KPOINTS ../resource/fp
mv ./in.lmp ../resource/md
mv ../OUTCAR ../resource/init_dt
cp ./1-convert_outcar_xsf.py ../resource/init_dt
cp ./2-convert_POSCAR_data.py ../resource/fp
cp ./3-makepot.py ../resource/fp
cd ../resource/init_dt/
python 1-convert_outcar_xsf.py
wait
rm OUTCAR
mv eval-set/* ./
cd ../fp
python 2-convert_POSCAR_data.py POSCAR data.nso
wait
mv data.nso ../md
python 3-makepot.py
rm POSCAR
wait
if [ "$command_line"="slurm" ]; then
sed -i "s|^#SBATCH --job-name.*|#SBATCH --job-name=$vasp_label_duty_name|" sub.slurm
elif [ "$command_line" = "PBS" ]; then
sed -i "s|^#PBS -N|#PBS -N $vasp_label_duty_name|" sub.slurm
else
echo "Command line is not 'slurm' or 'PBS. Run another command."
fi
cd ../md
if [ "$command_line"="slurm" ]; then
sed -i "s|^#SBATCH --job-name.*|#SBATCH --job-name=$md_duty_name|" sub-md.sh
elif [ "$command_line" = "PBS" ]; then
sed -i "s|^#PBS -N|#PBS -N $md_duty_name|" sub-md.sh
else
echo "Command line is not 'slurm' or 'PBS. Run another command."
fi
cd ..
sed -i "9s|.*|remote_work_place=$vasp_folder|" iter_scheduling_remote.sh
sed -i "s|^remote_ip=.*|remote_ip=$vasp_ip|" iter_scheduling_remote.sh
sed -i "s|^remote_port=.*|remote_port=$vasp_port|" iter_scheduling_remote.sh
sed -i "s|^task_sys=.*|task_sys=$command_line|" iter_scheduling_remote.sh
sed -i "s|^device.*|device = cuda:$GPU_number|" inv2
1-
import os
import sys
import time
import scipy.stats as st
import numpy as np
from sklearn.neighbors import KernelDensity
from matplotlib import pyplot
def timer(func):
def wrapper(*args, **kwargs):
start_time = time.time()
result = func(*args, **kwargs)
end_time = time.time()
print(f"Time elapsed: {end_time - start_time:.4f} seconds")
return result
return wrapper
def find_files(directory):
"""
递归查找一个目录下所有叫OUTCAR的文件,并返回这些文件的完整路径
"""
outcar_files = [] # 保存所有叫OUTCAR的文件的路径
for root, dirs, files in os.walk(directory):
for file in files:
if file == "OUTCAR":
outcar_files.append(os.path.join(root, file))
for dir in dirs:
dir_outcar_files = find_files(os.path.join(root, dir))
outcar_files.extend(dir_outcar_files)
# 将子目录添加到 dirs 中,以便递归处理
dirs[:] = [d for d in dirs if os.path.isdir(os.path.join(root, d))]
outcar_files = list(set(outcar_files))
return outcar_files
@timer
def read_vasp_xdatcar(file_path):
f = open(file=file_path)
f.readline()
f.readline()
lat = []
pos = []
# lattice
for _ in range(3):
lat.append(f.readline())
ele = f.readline()
npele = f.readline()
cnt = 0
tmp = None
while True:
line = f.readline()
if "configuration" in line:
cnt += 1
if tmp is not None:
pos.append(tmp)
tmp = []
continue
if not line:
pos.append(tmp)
break
tmp.append(line)
for conf_i in range(len(pos)):
for atom_i in range(len(pos[conf_i])):
pos[conf_i][atom_i] = pos[conf_i][atom_i].split()
pos[conf_i] = np.array(pos[conf_i]).astype(float)
ele = ele.split()
npele = npele.split()
npele = [int(i) for i in npele]
for i in range(len(lat)):
lat[i] = lat[i].split()
lat = np.array(lat).astype(float)
return cnt, ele, npele, lat, pos
def topn(arr, n):
indices = np.argpartition(arr, -n)[-n:]
values = arr[indices]
sort_indices = np.argsort(values)[::-1]
return values[sort_indices], indices[sort_indices]
def split_alp_num(string):
for i in range(len(string)):
if string[i].isdigit():
return [string[:i]], int(string[i:])
def write2my(file_path, ene_i, lat_i, ele_i, coo_i, foc_i):
lat_i = lat_i.reshape(3, 3)
coo_i = coo_i.reshape(-1, 3)
foc_i = foc_i.reshape(-1, 3)
with open(file_path, 'w') as file:
file.write(f"# total energy = {ene_i} eV\n\n")
file.write("CRYSTAL\n")
file.write("PRIMVEC\n")
for j in lat_i:
for k in j:
file.write(f'{k:20.8f}')
file.write('\n')
file.write("PRIMCOORD\n")
file.write(f"{len(coo_i)} 1\n")
for j in range(len(coo_i)):
file.write(f'{ele_i[j]:2}')
# coo
for k in coo_i[j]:
file.write(f"{k:20.8f}")
# force
for k in foc_i[j]:
file.write(f"{k:20.8f}")
file.write("\n")
pass
def direct2pos(lat, pos, direction=True):
"""
lat = [[Ax, Ay, Az],
[Bx, By, Bz],
[Cx, Cy, Cz]]
Pos = [n, 3]
:return: direct = [n, 3]
if direction=True
"""
if direction:
return pos @ lat
else:
return pos @ np.linalg.inv(lat)
class MyDistrib(st.rv_continuous):
def __init__(self, prdf):
super().__init__()
self.prdf = prdf
def _pdf(self, x, *args):
return self.prdf.evaluate(x)
class RejectSamp:
def __init__(self, ocr_name: str):
self.ocr_name = ocr_name # {XDATCAR path: OUTCAR path}
self.rng_min = 0
self.rng_max = None
self.rng_grid = 10000
self.rng = None
self.rng_intv = None
self.pdf = None
self.sum_intv = 10
self.envelope = None
self.samples = None
pass
def sampling(self, num):
nframe, natom, ele, ene, lat, pos, foc = self._anls_ocr()
print(nframe)
print(natom)
# print(ele)
print(ene.shape)
print(lat.shape)
print(pos.shape)
print(foc.shape)
self.rng_max = nframe
self.rng = np.linspace(self.rng_min, self.rng_max, self.rng_grid)
self.rng_intv = self.rng[1] - self.rng[0]
sd = np.sum((pos[1:] - pos[:-1]) ** 2, (1, 2))
nintv = sd.size // self.sum_intv
sum_sd = sd[:nintv * self.sum_intv].reshape(nintv, self.sum_intv).sum(1)
sum_sd_body = np.copy(sum_sd)
sum_sd_body[:sum_sd.size // 20] = sum_sd.min()
topn_val, topn_idx = topn(sum_sd_body, 100)
x_train = topn_idx * self.sum_intv
distrib = KernelDensity(kernel="gaussian", bandwidth=int(0.003 * nframe)).fit(x_train[..., None])
self.pdf = lambda x: np.exp(distrib.score_samples(np.reshape(x, [-1, 1])))
self.envelope = self.pdf(self.rng).max()
cnt = 0
iters = 0
res = np.array([])
while cnt < num:
u = np.random.uniform(self.rng_min, self.rng_max, num - cnt)
y = np.random.uniform(0, self.envelope, num - cnt)
pd = self.pdf(u)
res = np.append(res, u[pd > y])
res = res.astype(int)
res = np.unique(res)
cnt = len(res)
iters += 1
print(f"{iters:<3}{cnt:6}")
print("Total iters: ", iters)
self.samples = res
return self.samples.size, natom, ele, ene[self.samples], \
lat[self.samples], pos[self.samples], foc[self.samples]
def plot(self):
fig, ax = pyplot.subplots(figsize=(10, 6))
ax.hist(self.samples, bins=int(self.samples.size / 8), density=True, alpha=0.5)
ax.plot(self.rng, self.pdf(self.rng), 'r-', lw=1)
ax.plot(self.samples, -0.01 * self.envelope - 0.03 * self.envelope * np.random.random(self.samples.size), "+k")
ax.set_ylim(-self.envelope * 0.05)
ax.set_title(self.ocr_name + " PDF")
return fig
@timer
def _anls_ocr(self):
pos_foc = []
ene = []
lat = []
ions_per = []
types = []
print(self.ocr_name)
with open(self.ocr_name, 'r') as file:
while True:
line = file.readline()
# eof break
if not line:
break
# ions per type
#if "POSCAR" in line:
# ions_per.append(line)
# continue
if "ions per type" in line:
ions_per.append(line)
if "VRHFIN" in line:
types.append(line)
# lat
if "BASIS-vectors" in line:
file.readline()
file.readline()
file.readline()
file.readline()
for i in range(3):
line = file.readline()
lat.append(line)
continue
# pos & force
if "TOTAL-FORCE (eV/Angst)" in line:
line = file.readline()
while True:
line = file.readline()
if not line:
break
if "------" in line:
break
pos_foc.append(line)
continue
# ene
if "energy without entropy" in line:
ene.append(line)
ene = np.array([i.split('=')[1].split()[0] for i in ene]).astype(np.float64)
# lat = np.array([i.split() for i in lat]).astype(np.float64)[:, :3].reshape(-1, 3, 3)
lat = np.array([[float(i[4:][:12]), float(i[4:][12:25]), float(i[4:][25:38])] for i in lat]).astype(np.float64)[:, :3].reshape(-1, 3, 3)
# ele pretreatment
types = [i.split('=')[1].split(':')[0].rstrip() for i in types]
ions_per = ions_per[0].split()[-4:]
ele = sum([[i] * int(j) for i, j in zip(types, ions_per)], [])
pos_foc = np.array([i.split() for i in pos_foc]).astype(np.float64)
nframe = len(ene)
natom = len(ele)
pos = pos_foc[:, :3][:nframe * natom].reshape(nframe, natom, 3)[:nframe]
foc = pos_foc[:, 3:][:nframe * natom].reshape(nframe, natom, 3)[:nframe]
lat = lat[:nframe]
return nframe, natom, ele, ene, lat, pos, foc
if __name__ == '__main__':
# ocr_files = find_files("/data/ljx/third/lizhao/LYSO/LYSO-src/old")
#训练结构路径
ocr_files = ["./OUTCAR"]
#ocr_files = ["/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/triple/1",
# "/work/home/liz/workspace/1-system-MD/3-K5YSi4O12/9-1-lammps/6-train-NNP/outcar/triple/2"]
#训练集输出路径
out_path = "./"
if not os.path.exists(out_path):
os.mkdir(out_path)
out_data_path = os.path.join(out_path, "eval-set")
if not os.path.exists(out_data_path):
os.mkdir(out_data_path)
#每个数据取多少点,大概相乘在10000附近
smp_per_ocr = 50
# mode = 'rand'
mode = 'kde'
# # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # #
# Be careful when changing the code below
print(f"total trajectories: {len(ocr_files)}")
if not os.path.exists(out_data_path):
os.mkdir(out_data_path)
xsf_tot_cnt = 0
if mode == 'kde':
for ocr_i in range(len(ocr_files)):
r = RejectSamp(ocr_files[ocr_i])
n_frame, n_atom, ele, ene, lat, pos, foc = r.sampling(smp_per_ocr)
fig = r.plot()
fig_path = os.path.join(out_path, str(ocr_i))
fig.savefig(fig_path)
for j in range(n_frame):
path_j = os.path.join(out_data_path, f'structure{str(smp_per_ocr * ocr_i + j).zfill(5)}.xsf')
ene_j = ene[j]
lat_j = lat[j]
ele_j = ele
coo_j = pos[j]
foc_j = foc[j]
write2my(path_j, ene_j, lat_j, ele_j, coo_j, foc_j)
xsf_tot_cnt += 1
2-
# convert VASP OUTCAT to lammps .data (atomic)
#
# usage:
# python "this file" OUTCAR.input data.output
#
# # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # # ###
import numpy as np
from sys import argv
def anls_poscar(file_name: str):
with open(file_name, 'r') as f:
lines = f.readlines()
bloom_factor = float(lines[1])
lat = np.array([i.split() for i in lines[2: 5]]).astype(float)
lat = bloom_factor * lat
ele_and_num = lines[5][:-1] + ":" + lines[6][:-1]
pos_d = None
if (lines[7][0] == 'd') or (lines[7][0] == 'D'):
# direct coord.
pos_d = np.array([i.split() for i in lines[8:]]).astype(float)
elif (lines[7][0] == 'c') or (lines[7][0] == 'C'):
# cartesian
pos_c = np.array([i.split() for i in lines[8:]]).astype(float)
pos_d = direct2pos(lat, pos_c, False)
return lat, pos_d, ele_and_num
def direct2pos(lat, pos, direction=True):
"""
lat = [[Ax, Ay, Az],
[Bx, By, Bz],
[Cx, Cy, Cz]]
Pos = [n, 3]
:return: if True: return cart = [n, 3]
if False: return direct = [n, 3]
"""
if direction:
return pos @ lat
else:
return pos @ np.linalg.inv(lat)
def clip_ny_norm(x):
return x / np.linalg.norm(x)
def cart2lams(lat):
"""
vasp (POSCAR) Coordinate System:
O = [[ax, ay, az],
[bx, by, bz],
[cx, cy, cz]]
:return:
lammps Coordinate System:
O' = [[xhi - xlo, 0, 0],
[xy, yhi - ylo, 0],
[xz, yz, zhi - zlo]]
"""
ax = (lat[0] ** 2).sum() ** 0.5
a_hat = clip_ny_norm(lat[0])
bx = (lat[1] * a_hat).sum()
by = np.linalg.norm(np.cross(a_hat, lat[1]))
cx = (lat[2] * a_hat).sum()
cy = (lat[2] * np.cross(clip_ny_norm(np.cross(lat[0], lat[1])), a_hat)).sum()
cz = np.abs((lat[2] * clip_ny_norm(np.cross(lat[0], lat[1]))).sum())
new = np.array([
[ax, 0, 0],
[bx, by, 0],
[cx, cy, cz]
])
return new
def write_lmp_atomic_data(filename, lmp_lat, pos_c, strr):
new_coo = pos_c
Z_of_type = strr.split(':')[0].split()
n_type = strr.split(':')[1].split()
new_ele = [[i] * int(n_type[i - 1]) for i in range(1, len(n_type) + 1)]
new_ele = sum(new_ele, [])
# write
with open(filename, "w") as f:
f.writelines("%s (written by ARES-NNP)\n\n" % str(filename + strr))
f.writelines("%i atoms\n" % len(new_coo))
f.writelines("%i atom types\n" % len(Z_of_type))
f.writelines("0.0 %.17f xlo xhi\n" % lmp_lat[0][0])
f.writelines("0.0 %.17f ylo yhi\n" % lmp_lat[1][1])
f.writelines("0.0 %.17f zlo zhi\n" % lmp_lat[2][2])
f.writelines(
" %18.12f %18.12f %18.12f xy xz yz\n\n\n" % (lmp_lat[1][0], lmp_lat[2][0], lmp_lat[2][1]))
f.writelines("Atoms\n\n")
index_list = np.arange(1, len(new_coo) + 1)
for i, ele, coo in zip(index_list, new_ele, new_coo):
f.writelines("{0:>6} {1:>3} {2:20.12f} {3:20.12f} {4:20.12f}\n".format(i, ele, coo[0], coo[1], coo[2]))
if __name__ == '__main__':
filename = argv[2]
lat, pos, strr = anls_poscar(argv[1])
crs_para = np.linalg.norm(lat, axis=1)
exg = np.argsort(crs_para)[::-1]
lat = lat[exg, :]
pos = pos[:, exg]
lmp_lat = cart2lams(lat)
inv = np.linalg.inv(lat) @ lmp_lat
pos_c = direct2pos(lat, pos, True) @ inv
write_lmp_atomic_data(filename, lmp_lat, pos_c, strr)
3-
# 字典:元素对应的赝势文件路径
base_pseudo_path = "/public/lijx/work/Na-X-Si-O/pot-makepot/potpaw_PBE/"
pseudo_paths = {
'H': 'H/POTCAR',
'Li': 'Li_sv/POTCAR',
'Be': 'Be_sv/POTCAR',
'B': 'B/POTCAR',
'C': 'C/POTCAR',
'N': 'N/POTCAR',
'O': 'O/POTCAR',
'F': 'F/POTCAR',
'Ne': 'Ne/POTCAR',
'Na': 'Na_pv/POTCAR',
'Mg': 'Mg_pv/POTCAR',
'Al': 'Al/POTCAR',
'Si': 'Si/POTCAR',
'P': 'P/POTCAR',
'S': 'S/POTCAR',
'Cl': 'Cl/POTCAR',
'K': 'K_sv/POTCAR',
'Ca': 'Ca_sv/POTCAR',
'Sc': 'Sc_sv/POTCAR',
'Ti': 'Ti_pv/POTCAR',
'V': 'V_pv/POTCAR',
'Cr': 'Cr_pv/POTCAR',
'Mn': 'Mn_pv/POTCAR',
'Fe': 'Fe_pv/POTCAR',
'Co': 'Co/POTCAR',
'Ni': 'Ni_pv/POTCAR',
'Cu': 'Cu_pv/POTCAR',
'Zn': 'Zn/POTCAR',
'Ga': 'Ga_d/POTCAR',
'Ge': 'Ge_d/POTCAR',
'As': 'As/POTCAR',
'Se': 'Se/POTCAR',
'Br': 'Br/POTCAR',
'Kr': 'Kr/POTCAR',
'Rb': 'Rb_sv/POTCAR',
'Sr': 'Sr_sv/POTCAR',
'Y': 'Y_sv/POTCAR',
'Zr': 'Zr_sv/POTCAR',
'Nb': 'Nb_pv/POTCAR',
'Mo': 'Mo_pv/POTCAR',
'Tc': 'Tc_pv/POTCAR',
'Ru': 'Ru_pv/POTCAR',
'Rh': 'Rh_pv/POTCAR',
'Pd': 'Pd/POTCAR',
'Ag': 'Ag/POTCAR',
'Cd': 'Cd/POTCAR',
'In': 'In_d/POTCAR',
'Sn': 'Sn_d/POTCAR',
'Sb': 'Sb/POTCAR',
'Te': 'Te/POTCAR',
'I': 'I/POTCAR',
'Xe': 'Xe/POTCAR',
'Cs': 'Cs_sv/POTCAR',
'Ba': 'Ba_sv/POTCAR',
'La': 'La/POTCAR',
'Ce': 'Ce_3/POTCAR',
'Pr': 'Pr_3/POTCAR',
'Nd': 'Nd_3/POTCAR',
'Pm': 'Pm_3/POTCAR',
'Sm': 'Sm_3/POTCAR',
'Eu': 'Eu_3/POTCAR',
'Gd': 'Gd_3/POTCAR',
'Tb': 'Tb_3/POTCAR',
'Dy': 'Dy_3/POTCAR',
'Ho': 'Ho_3/POTCAR',
'Er': 'Er_3/POTCAR',
'Tm': 'Tm_3/POTCAR',
'Yb': 'Yb_2/POTCAR',
'Lu': 'Lu_3/POTCAR',
'Hf': 'Hf_pv/POTCAR',
'Ta': 'Ta_pv/POTCAR',
'W': 'W_pv/POTCAR',
'Re': 'Re_pv/POTCAR',
'Os': 'Os_pv/POTCAR',
'Ir': 'Ir/POTCAR',
'Pt': 'Pt/POTCAR',
'Au': 'Au/POTCAR',
'Hg': 'Hg/POTCAR',
'Tl': 'Tl_d/POTCAR',
'Pb': 'Pb_d/POTCAR',
'Bi': 'Bi/POTCAR',
'Th': 'Th/POTCAR',
'Pa': 'Pa/POTCAR',
'U': 'U/POTCAR',
'Np': 'Np/POTCAR',
'Pu': 'Pu/POTCAR',
# 添加其他元素和对应的赝势路径
}
for element, relative_path in pseudo_paths.items():
full_path = f"{base_pseudo_path}{relative_path}"
pseudo_paths[element] = full_path
# 读取 POSCAR 文件
def read_poscar(poscar_path):
with open(poscar_path, 'r') as f:
lines = f.readlines()
elements = lines[5].split()
periodic_table_order = ['H', 'He', 'Li', 'Be', 'B', 'C', 'N', 'O', 'F', 'Ne','Na', 'Mg', 'Al', 'Si', 'P', 'S', 'Cl', 'Ar','K','Ca', 'Sc', 'Ti', 'V', 'Cr', 'Mn', 'Fe', 'Co','Ni', 'Cu', 'Zn','Ga', 'Ge', 'As', 'Se', 'Br', 'Kr', 'Rb', 'Sr', 'Y', 'Zr', 'Nb','Mo', 'Tc', 'Ru', 'Rh', 'Pd', 'Ag', 'Cd', 'In', 'Sn', 'Sb', 'Te','I', 'Xe', 'Cs', 'Ba', 'La', 'Ce', 'Pr', 'Nd', 'Pm', 'Sm', 'Eu','Gd', 'Tb', 'Dy', 'Ho', 'Er', 'Tm', 'Yb', 'Lu', 'Hf', 'Ta', 'W','Re', 'Os', 'Ir', 'Pt', 'Au', 'Hg', 'Tl', 'Pb', 'Bi', 'Po','At','Rn','Fr','Ra','Ac','Th', 'Pa','U', 'Np', 'Pu', 'Am', 'Cm', 'Bk', 'Cf', 'Es', 'Fm', 'Md', 'No', 'Lr']
sorted_elements = sorted(elements, key=lambda x: periodic_table_order.index(x))
return sorted_elements
# 创建大的 POTCAR 文件
def create_big_potcar(elements, pseudo_paths, output_path):
with open(output_path, 'w') as f_out:
for element in elements:
if element in pseudo_paths:
pseudo_path = pseudo_paths[element]
with open(pseudo_path, 'r') as f_pseudo:
f_out.write(f_pseudo.read())
else:
print(f"赝势文件不存在或未定义:{element}")
def copypot(elments,pseudo_paths):
import os
for element in elements:
output_path2 = "POT-"+element
if element in pseudo_paths:
pseudo_path = pseudo_paths[element]
with open(output_path2, 'w') as f_out:
with open(pseudo_path, 'r') as f_pseudo:
f_out.write(f_pseudo.read())
else:
print(f"赝势文件不存在或未定义:{element}")
if __name__ == "__main__":
poscar_path = "POSCAR" # 输入文件名
output_potcar_path = "POTCAR" # 输出的大 POTCAR 文件名
elements = read_poscar(poscar_path)
create_big_potcar(elements, pseudo_paths, output_potcar_path)
copypot(elements,pseudo_paths)
print("大的 POTCAR 文件已创建")
nohup ./scheduling.sh > log 2>&1 &
3. 华为新版本
1.变化
增加了server.sh
改变了iter_scheduling_remote.sh 现在,这两个都不需要做改变可以直接复制

2. 技巧
1.各个文件夹含义
pre-select :主动学习最后一步训练inv2里用的是train_dt,之前步用的看上去是pre-select,但实际上运行时用的也是train_dt
train_dt
非主动学习训练:只需要把主动学习inv2的restart改为false,不能删掉,记得更改inv2中cuda的编号,只申请了一张卡
如果一直训练出的只能跑几步,可以尝试修改select_thr = 2.0 为1.2
4. 主动学习能量、力误差
主动学习训练势过程中要求能量和力的误差不能太大,因此需要注意监控能量、力的误差
存在的一种可能是,主动学习的能量和力误差太大,但是可以正常跑完主动学习的过程(如111胞 1ns)。这时候,由于主动学习的误差很大,因此不能认为跑出的1ns结果是合理的。
目前的版本中还不能设置在能量和力达到一个标准时自动停止,因此需要多注意能量和力
提升训练精度的方法
- 学习率和训练圈数是重要的可设置的参数,通过增加训练圈数,可以有效地提高训练的精度(降低能量和力的误差)
- 清洗数据,可能存在某个误差值很大的结构,导致整体训练效果变差,把这个结构删除掉,能够有效降低误差
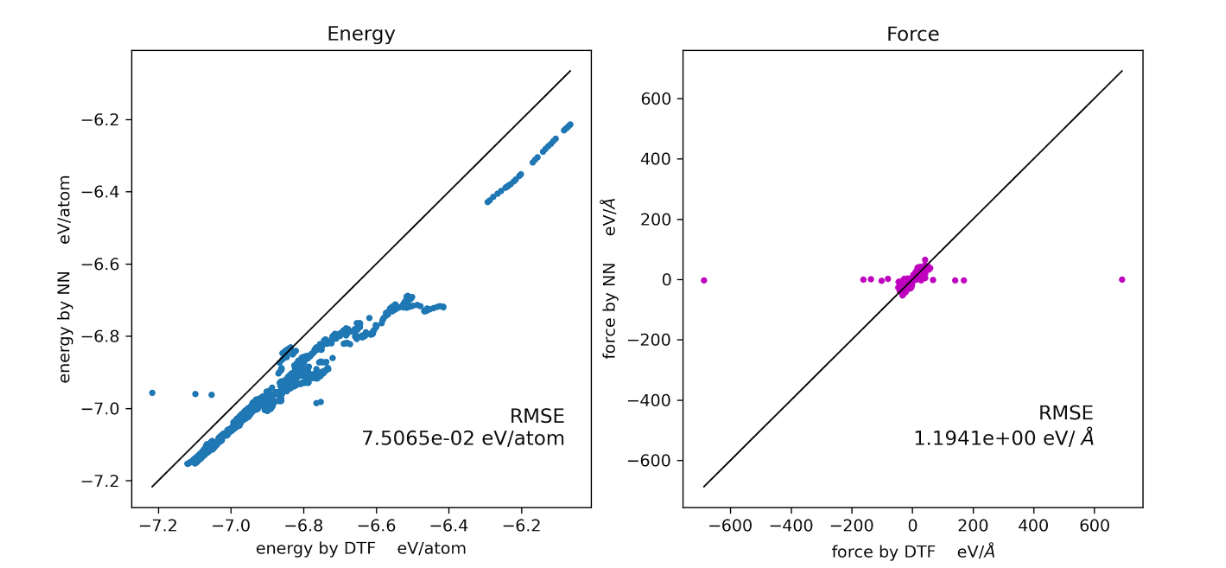
5. 主动学习+ minima_hopping
1. minima_hopping使用方法
minimaohopping是在ase中使用的一种采样算法,相比普通的md,采样更有效,这在华为机器liz/2-Na-X-Si-O/2_NaXSiO_big_model/1_one_element/05_Sb_mini 中得到了充分的验证。
使用minimahopping需改动的地方:
resource/md文件夹中的data.nso 改为data.vasp,data.vasp的文件内容格式为POSCARresource/md文件夹中增加minima_hopping.py,用于进行minima_hopping模拟,可直接copy,需要修改脚本内的元素类型resource/md文件夹中的sub-md.sh需要改变,从提交md变为提交miniresource文件夹中的iter_scheduling_remote.sh 有一点改变,可直接copy
sub-md.sh,iter_scheduling_remote.sh, mimima_hopping.py 可直接从华为机器/liz/2-Na-X-Si-O/2_NaXSiO_big_model/1_one_element/05_Sb_mini/resource 路径下复制到对应位置,minima_hopping.py需要修改元素类型
6. 经验总结
关于训练集
- 需要用高精度的第一性原理数据来进行势的训练
- 结构优化过程中的结构不能用来进行势的训练,因为计算的能量不准确
- 不同ENCUT和k点密度的结构不能用来训练同一个势
- 升温过程
- 增加压力点
- NPT系综
- meta-dynamics
- minim
- AIMD加大步长
- 添加拉伸剪切数据集
关于训练一个效果好的势
- 机器学习计算高温比低温更难,因此高温的数据集很重要
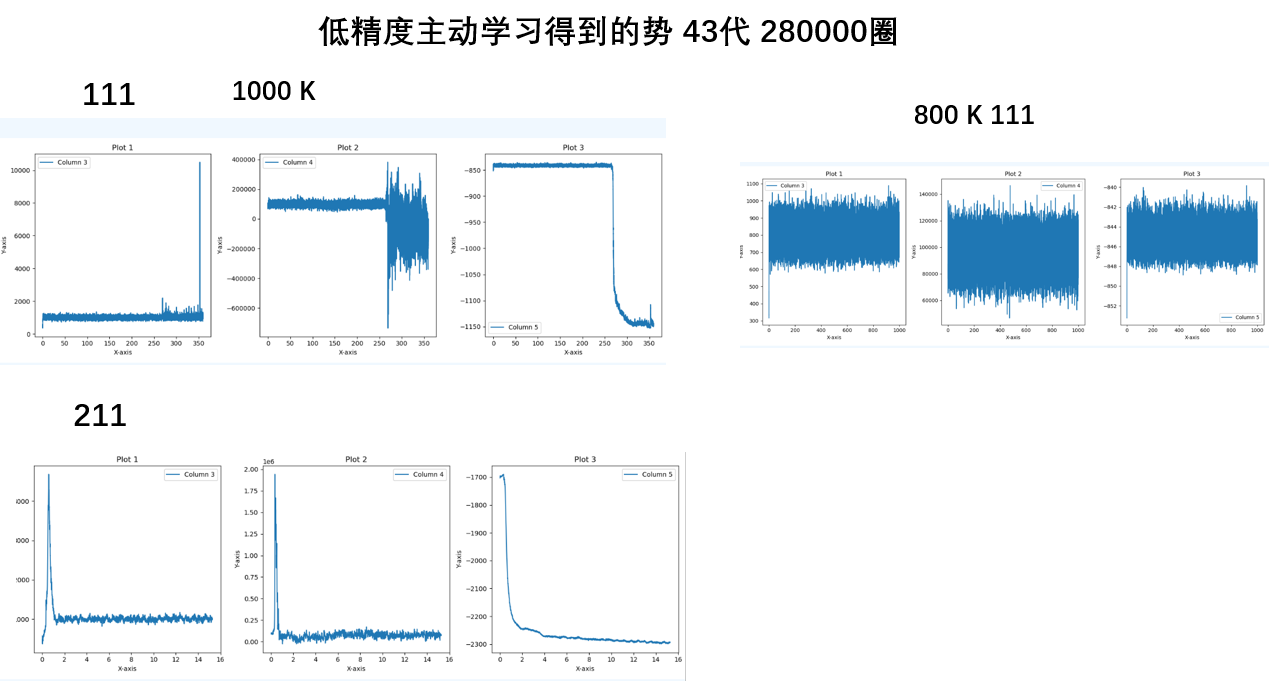
- 训练圈数多的要好于少,尽管大小圈数的精度可能相同
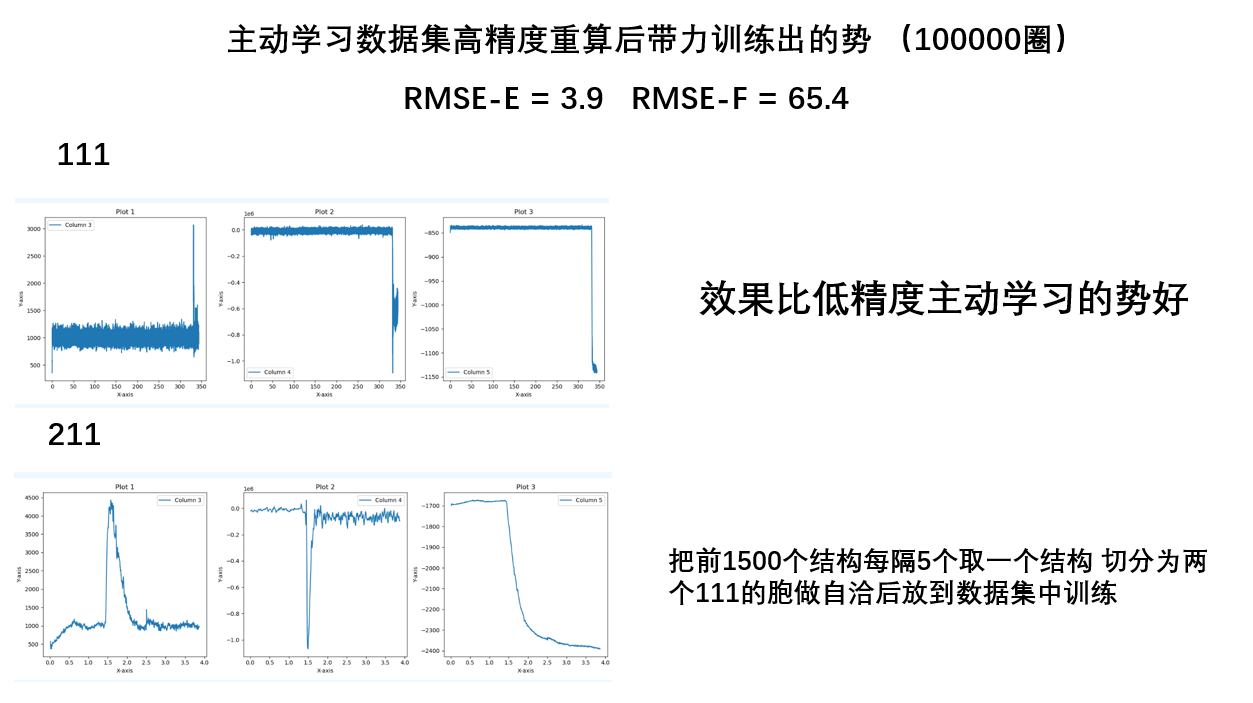
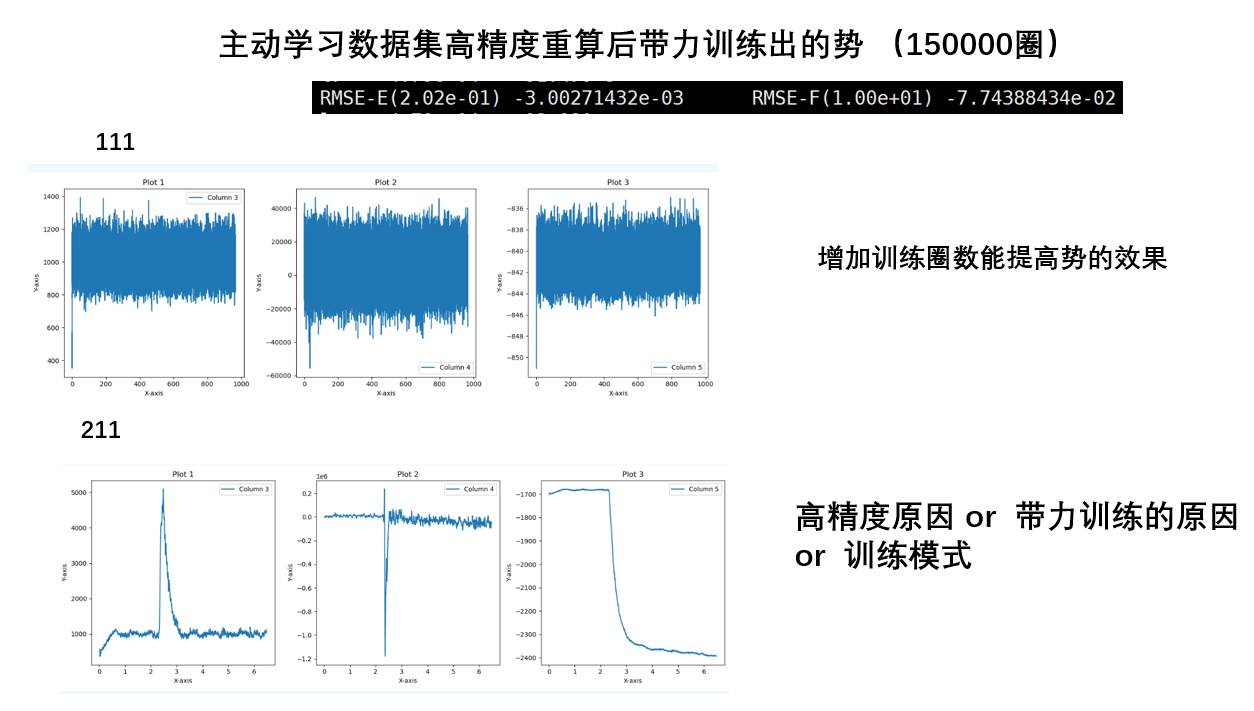
- 高精度数据集比低精度好,高精度的数据集训练的势更好
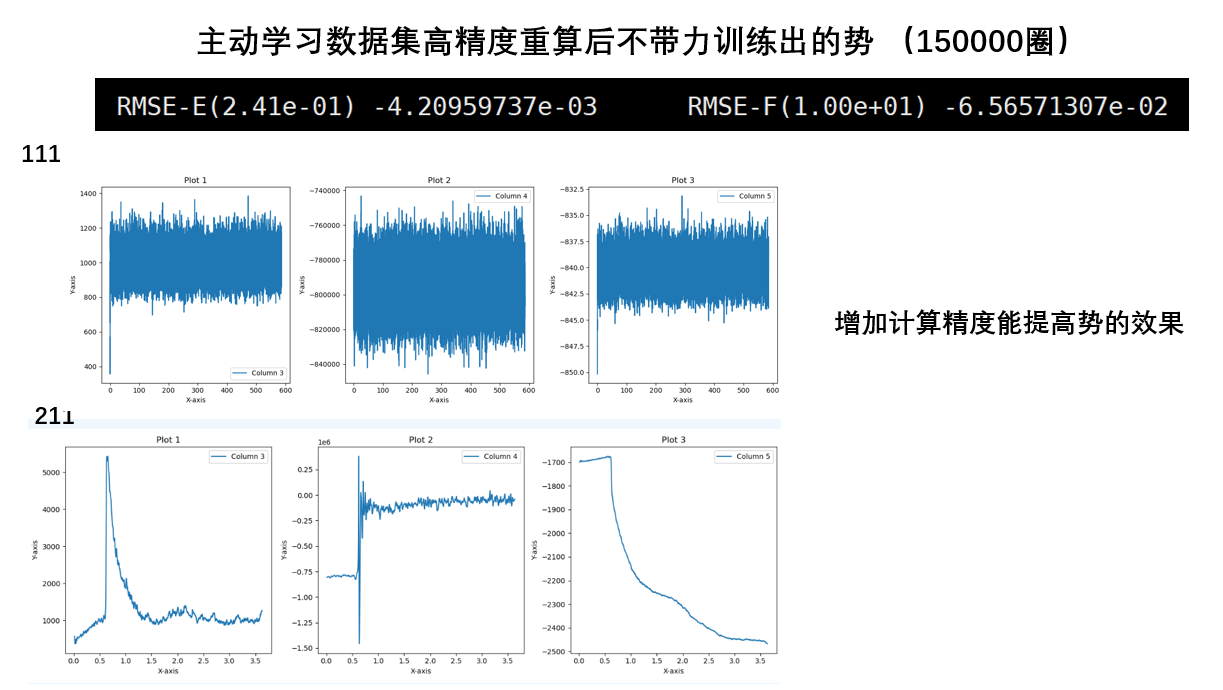
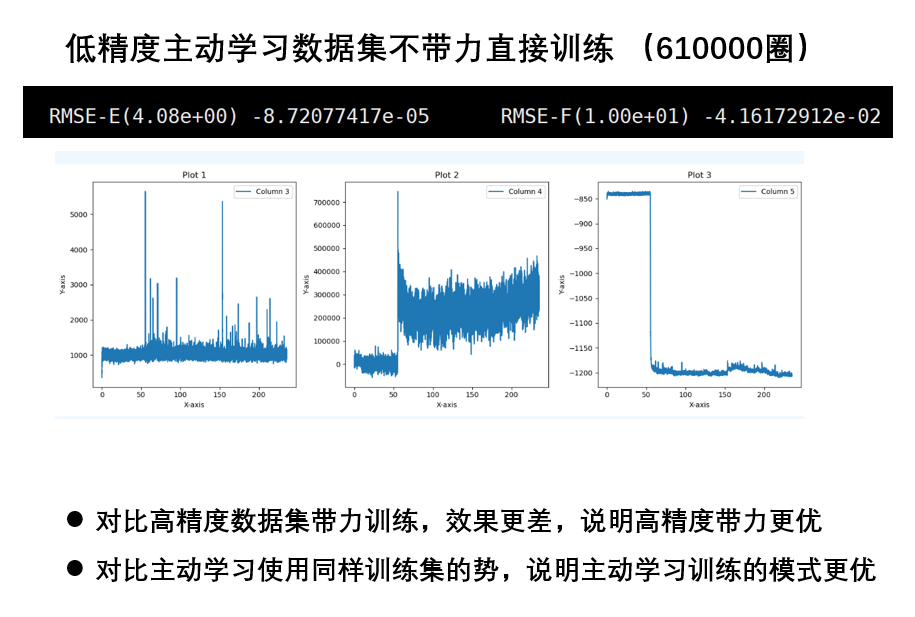
- 主动学习的势要比普通训练更优
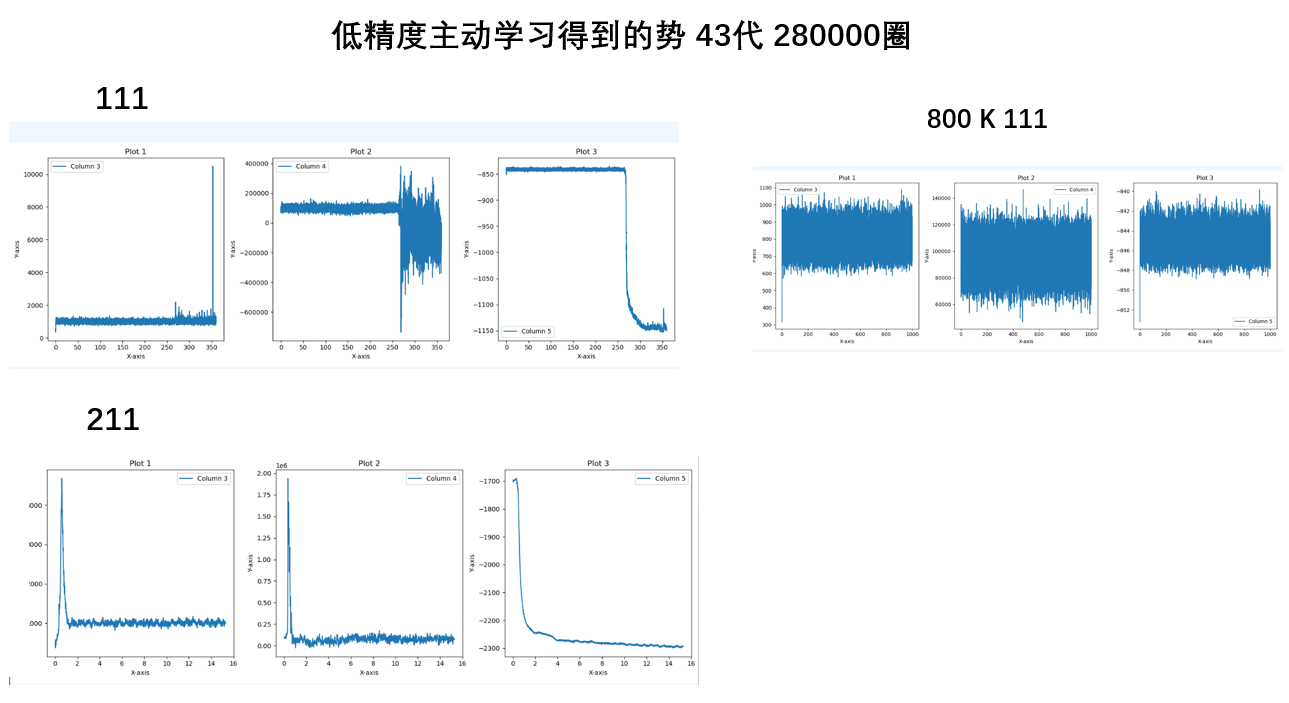
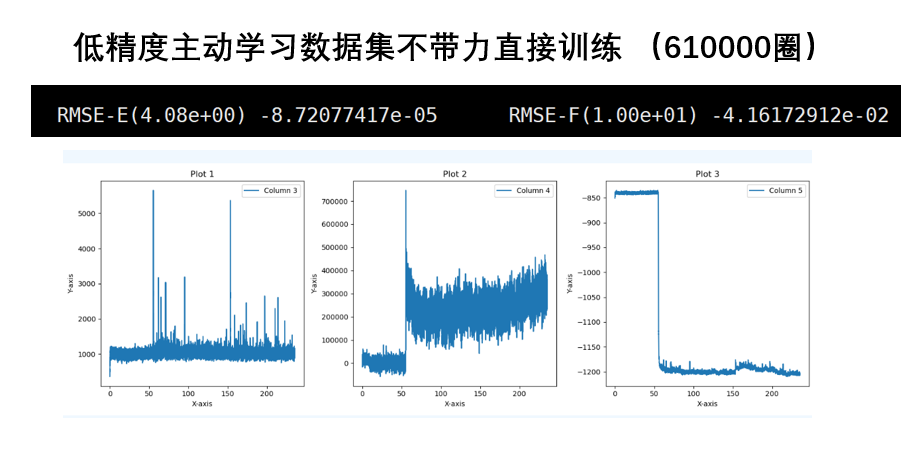
- 如果不算压强,带不带力的计算结果变化并不大
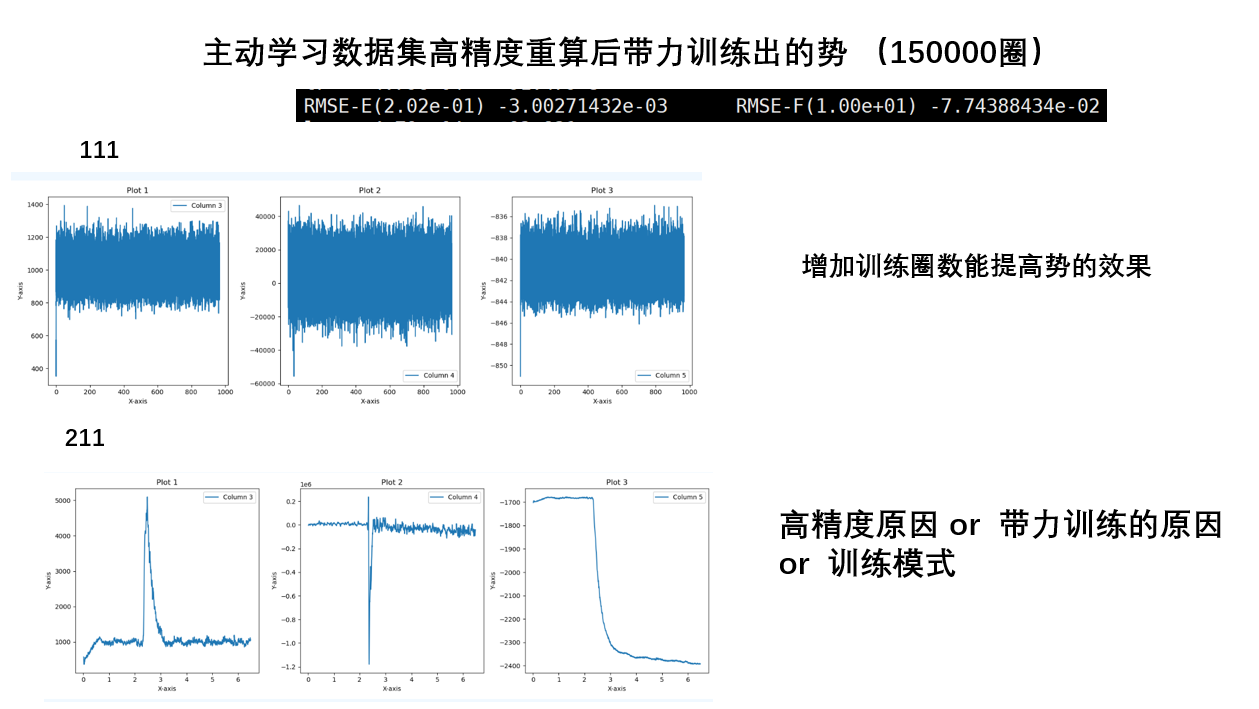
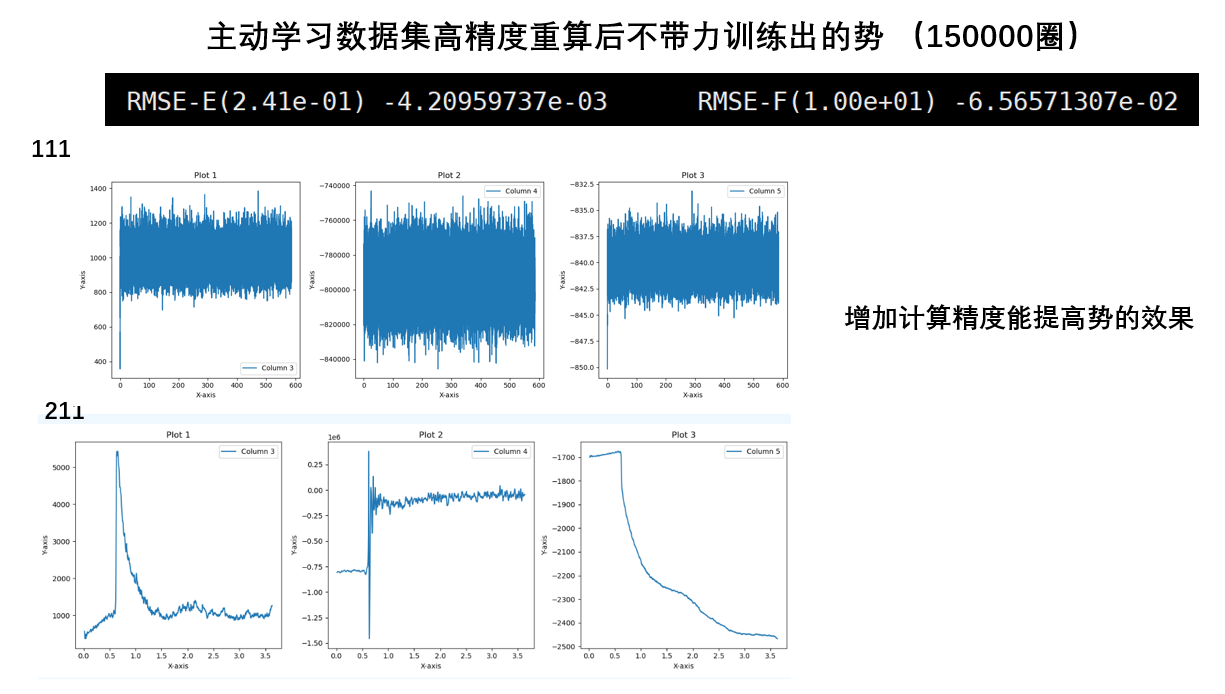
关于训练势的误差
1.
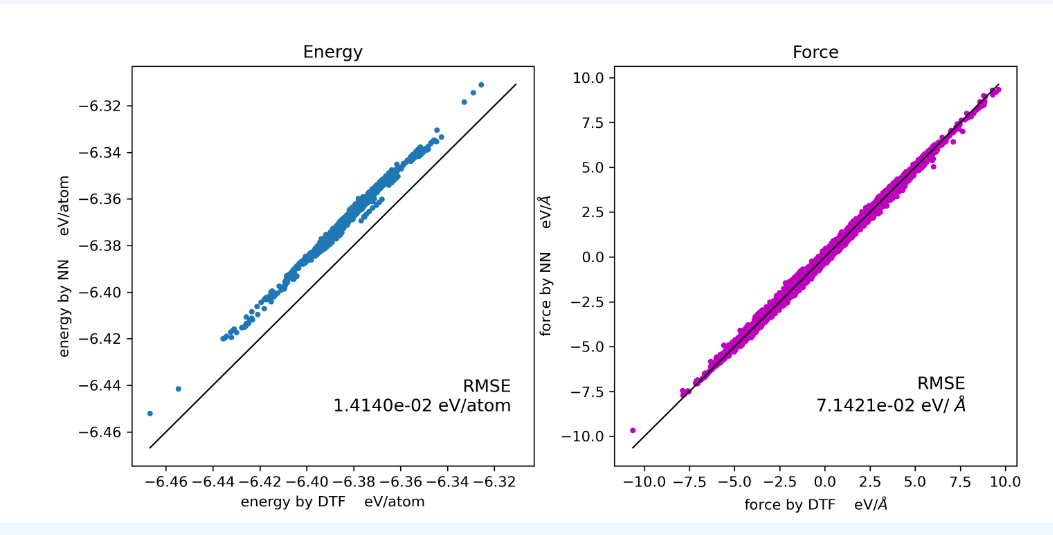
能量的误差不在线上是没问题的,因为机器学习框架中会加一个常数项,现在学习率还是太高,导致这个常数项比较高。通过降低学习率可以解决这个问题(手动降低或者训练足够长的时间自动降低)
- 一个结构对应着1个能量,但是对应着3*原子数个力,所以一个结构的能量偏差大,对应的是许多个力的偏差大
7. 代码理解
Ap_*由sub-select根据现有的结构产生,和训练势过程和分子动力学过程无关。
主动学习和普通机器学习的区别
- 训练势时是从上一个势开始训练,增加新的数据集,重置学习率等
- 跑分子动力学时in.lmp添加al,并且使用由select.sh生成的Ap_*,用来监控并终止分子动力学,产生oos
sub-select.sh需要在GPU上跑,不然会报错

acnn -grade 用Ap_*产生某一个结构的grade,如果grade值高于某一个数,就可以加到oos中
主动学习停掉的情况
- grade值大于3且小于八累计了50个结构
- grade值大于8
主动学习的两次select
第一次是md过程中select不能判断的结构,第二次select是产生oos后对相似oos结构的一个去重,然后选择grade值相对大一些的结构(张开一个maxvol空间,选取边缘)
第一次是用现有的Ap_inv ,第二次是新产生一批Ap_inv
转载请注明来源 有问题可通过github提交issue